Resurrecting the ancient Israelites from the Valley of Dry Bones
by geneticist Dr. Eran Elhaik
[Editor’s Note: VT has known Israeli geneticist Eran Elhaik since 2013 when we spotted the Jewish genome work he was doing at Johns Hopkins.

We noted right away that he was another of the many academics who do not let ideology and politics influence his scientific research, which is always a breath of fresh air in the geopolitical sewer that VT has to swim in every day.
His work was politically controversial but solid, and he was able to defend it from the shills that attempted to discredit it.
In short, his work showed that Ashkenazic Jews have about as much “people of the land” DNA as some Europeans have Neanderthal DNA. But Europeans have never referred to themselves as ancient Neanderthals nor laid claim to Africa as the “ancient homeland”, other than the former colonialists before conditions drove them into exile.
Here is my first article I wrote on Elhaik’s work, New Genome Study Destroys Zionist Claims to Palestine, published in the Press TV Viewpoints commentary section in the heydays when VT writers were constantly in the top five read articles each week.

In the past six years, new Neolithic remains discoveries in Israel and the advances in genetic sequencing have combined to support Elhaik’s 2013 work, that Ashkenazi Israelis tried to pull off what he aptly described as “shooting the arrow and then drawing a bullseye around it.”
His work made a deep impression on Gordon and me, how this one man with his scientific integrity stood up to the fake Hollywood Ashkenazic Zionist claim to the ancient land. A year after that, five years ago, VT began writing about fake news, how it was being done on an industrial scale, not just a few bad apples.
We were never attacked over our claims but were ignored. And then magically, about two years ago, the term “fake news” began being widely used.
Elhaik describes below that the Yemenis have the closest blood links to the ancient Israelis, while the Saudis, with support from Israel, have actually been doing a mini-genocide on these closest relatives of the Ancients.
Do take a look at Elhaik’s new Primeval DNA test, which goes way beyond letting us know we are all descended from early Africans. Most of us are more interested in where our ancestors were a few thousand to 10K years ago, and his new project can compare your DNA to the pool of ancient remains that have had their DNA sequenced … Jim W. Dean ]
The hand of the LORD was on me, and he brought me out by the Spirit of the LORD and set me in the middle of a valley; it was full of bones. He led me back and forth among them, and I saw a great many bones on the floor of the valley, bones that were very dry
And as I was prophesying, there was a noise, a rattling sound, and the bones came together, bone to bone.
I looked, and tendons and flesh appeared on them and skin covered them, but there was no breath in them… Then he said to me: “Son of man, these bones are the people of Israel.” ~ Ezekiel 37

– First published … May 11, 2019 –
Dr. Eran Elhaik is a population, medical, and evolutionary geneticist at the University of Sheffield, UK. Eranelhaiklab.org. Reprinted on VT by permission of the author.
The Vision of the Valley of Dry Bones prophecy is one of the most powerful prophecies of the prophet Ezekiel. In this vision, Ezekiel finds himself in a valley full of dry human bones of Israelite origins. He is asked to revive them, bring them hope, and lead them to the land of Israel.
For the past decades, Paleogenomics, the field dealing with ancient genomes, has been fulfilling Ezekiel’s vision by extracting DNA from dry bones and telling their stories with increasing accuracy.
As Ezekiel envisioned, only dry (and cold) bones can tell stories. Humidity and heat accelerate the degradation of the DNA beyond repair. At times, even the samples we can use are of poor quality, leading to an inability to put together all the pieces of the DNA and thus incomplete stories from the bones as if told by stuttering tongueless skeletons. These are only some of the challenges we face in our efforts to reconstruct the past.
Ezekiel’s prophecies were written in the 6th century after several exiles of the Judeans to Babylon (601-582 BC). The importance of this vision to the debate on the continuity of the Jewish people became apparent very quickly. Already in the Talmud, two interpretations were offered. The first considered it a complete allegory.
The second considered it a reality where the Babylonian exiles returned to Israel and continued the Judaean bloodline. Zionism, a secular movement that recruited the Bible to serve its needs whenever necessary, considered the State of Israel the ultimate fulfillment of Ezekiel’s prophecies and called to the ingathering of the exiles.
The question of whether Ezekiel’s vision has been correctly interpreted is not anachronistic and has offshoots in history, archeology, linguistics, and genetics – the battlefields of the primordialists and constructivists.
Did contemporary Jews descend from the ancient Israelites, as the primordialists claim? Or did they descend from people who converted to Judaism and only later on adopted nationalist ideas as the constructivist camp argues?
Of course, one look at Ethiopian, Yemenite, Ashkenazic, and Russian Jews can lend credence to the constructivist view without digging up a single bone. However, the primordialist camp has ready-made answers to that question. It criticized the weak diasporas Jews who succumbed to assimilation and embraced the Jews who remained as authentic Israelites.
It has always been clear that the only way to decide which Jewish communities represent the ancient Israelites most accurately, we should go back to the source – Israel. Therefore, in the late 19th century a search for the jüdische Typus, the “Jewish type” was launched. Anthropologists explored Palestine, studied the native inhabitants, and compare their anthropological measures to Jews.
The results of these studies were incredibly disappointing, at least to the Ashkenazic Jews who carried out these studies and craved to see the jüdische Typus reflected in the mirror. As it turned out, Yemenite Jews held the greatest anthropological resemblance to the wandering Bedouins, the poster boys of the Patriarchs.
Ashkenazic Jews, by contrast, resembled the Caucasian type more than anything else (Efron 1994). This was not what Zionist leaders wanted to hear amid the formation of their nationalistic movement that called Jews to return to their homeland, fight, and, maybe, die for it. The research question had to be rephrased.
Abandoning any desire to see a jüdische Typus specimen ever again, the new research paradigm focused on studying features common to all contemporary Jews and deriving the characteristics of the jüdische Typus from them (Elhaik 2016). Unfortunately, no biomarker for Jewishness was ever found in a way that excluded Jews from non-Jews.
Yet the question remained: which of all the Jews who have nothing in common except religion, best represent the Ancient Israelites.
The answer among the Ashkenazic Jewish researchers was almost unanimous – let us mold the jüdische Typus in our own image.
To support these claims, geneticists began producing a large body of literature aimed to support and prove two things:
- their genetic superiority and
- their genetic ties to Israel, by showing their resemblance to Levantine populations (Falk 2017), whose own claims to the land were later dismissed on account of being “work migrants.” Kirsh (2003) demonstrated how human geneticists and physicians have consistently manipulated their results and emphasized the sociological and historical aspects of their research using their work as a vehicle for establishing a national identity and confirming the Zionist narrative.
The mammoth in the room was the lack of any genetic evidence from the ancient Israelites that would allow testing the similarity of their DNA to that of modern-day Jews. Since no one imagined that mammoths would ever come to life, they avoided the problem completely.
It was much easier to pretend that modern-day Jews and, Ashkenazic Jews, in particular, are living replicas of the ancient Israelites who not only were all related to one another but also resisted the gene flow from non-Jews all this time.
One such enthusiastic advocate is Harry Ostrer (2012), who incidentally tipped his hand by explaining, “The stakes in the genetic analysis are high. It is more than an issue of who belongs in the family and can partake in Jewish life and Israeli citizenship.
It touches on the heart of Zionist claims for a Jewish homeland in Israel. One can imagine future disputes about exactly how large the shared Middle Eastern ancestry of Jewish groups has to be to justify Zionist claims.”
It seems that Ostrer, a resident of a former Native Indian land now known as New York, is so passionate to gain rights to more lands that he completely forgot that his recent 2010 study (Atzmon et al. 2010) ascribed only 50% of Middle Eastern ancestry to Jews compared to 56-59% of non-Jews.
Ostrer’s proposal, thereby, grants non-Jews the largest proportion of the land although they represent only 25% of the general population. One has to assume that those 50% Middle Eastern ancestry of Jews must have experienced mitosis between the two publications to avoid considering Dr. Ostrer’s suggestions as discriminatory toward the Jewish people.
Despite the imagined link between modern-day Jews and the ancient Israelites, the claims of the primordialist camp became well accepted in the direct-to-consumer industry.
The myth of the “Cohen Gene” (Skorecki et al. 1997) or “Four mitochondrial mothers” (Behar et al. 2004) produced by the members of this camp became the bedrock of the Genetic Judaism era, where one needs only to order a genetic test from the right company to receive a Jewishness certificate.

Paleogenomics changed all that. Thanks to advances in the field, it became possible to extract DNA from ancient people and identify their mitochondrial haplogroups and even autosomal DNA.
This remarkable progress allowed the unthinkable: A semi-revival of the ancient Israelites from their dry bones and the recovery of their stories. The DNA extracted from those bones can tell us who these people were, what they looked like, what they ate, and what diseases they carried (Nielsen et al. 2017; Prohaska et al. 2019).
We can trace their migration routes to gain a deeper understanding of where we all came from. Yes, they can also tell us how similar those ancient Israelites were to modern-day people, and if modern-day Jews are not the lineal descendants of the ancient Israelites, we can find out who are.
To answer some of these questions, I launched the Primeval DNA test.
The test allows anyone to upload their autosomal DNA file obtained from any genetic company and compare themselves to ancient people from an initial set of seven populations: Ice Age Europeans, Stone Age Europeans, Medieval Vikings, Egyptian mummies, Roman Britons, Chumash Paleo-Indians, and Ancient Israelites.

The ancient Israelites were obtained from three regions Motza Tachtit at the territory of the tribe of Benjamin, Peqi’in (Naphtali), and Rakefet Cave (Manasseh), near the valley of the Rakefet river.
The results can be discussed in our FB group. I also launched the Ancient DNA Hub as a source of knowledge on ancient cultures whose DNA has have been sequenced.
The Primeval DNA test is the most potent genetic tool publicly available to test claims of ancient origins. It allows anyone to journey to their deep past, and explore for the first time their genetic similarity with ancient people. The bones still cannot speak, but in their own way, they have a lot to teach us and allow us to reevaluate our core beliefs.
The answer to the question of who is closer to the ancient Israelites rests in the DNA extracted from some 50 bones of ancient Israelites and Judaeans with many more to come. Are modern-Jews mostly Middle Easterns (or ancient Israelites) like Ostrer claims? Hardly.
I ran the test on 80 Jews from various communities. Upon examining the results, I could not help noticing the irony considering how the progress in population genetics validated the findings of the 19th-century anthropologists who combed the Levant in an honest search for the jüdische Typus before the repercussions of their findings became clear and their results were deplored on every stage.
The most similar Jews to the ancient Israelites who left their dry bones in the Rakefet Valley in Israel were Yemenite Jews, but that genetic similarity was less than 10%, on average, in agreement with our previous analyses ascribing less than 5% ancient Levantine ancestry to Ashkenazic Jews (Das et al. 2017) and in support of their potential relatedness to the Ashina tribe.
Yet, these averages mask the high heterogeneity among all Jewish communities. Some people may share the highest similarity with Gal (named after Wonder Woman actress Gal Gadot), a young Neolithic woman – only 6200 years old, and other people may find that they are close to Abraham, a Turkish man (E1b1) who led a group of Anatolians to what he must to have felt was the promised land.
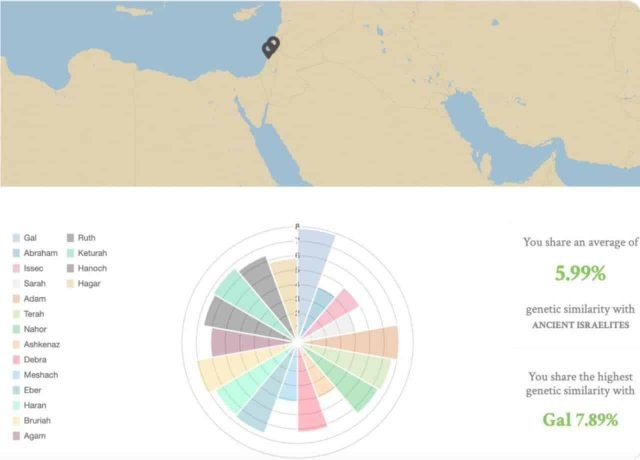
Due to the many population replacements that the area experienced, we can see a diverse range of mitochondrial haplogroups that vary over time. Among the most common lineages are J2, K1a, and T. An analysis of Judaeans from the first century AD confirmed the prevalence of the T haplogroup (Matheson et al. 2009), found today in less than 10% of Ashkenazic Jews.
Unsurprisingly, not a single skeleton matches the alleged four Ashkenazic Jewish mothers, whose origin is in prehistoric Europe (Costa et al. 2013). As expected, an exact match with one of those mothers was found in Neolithic Spain (Haak et al. 2015). Interestingly, the Y chromosomal haplotypes of the ancient Israelites are typically E1b1 and T1 haplotypes, commonly found today in Africa with lower frequencies in the Middle East and Europe.
We can expect that future tests covering other regions of the world would be able to explain the remaining portion of the elusive Jewish ancestry. Only time will say if “Genetic Jewishness” will evolve onto “Primeval Jewishness” where people define their Jewishness based on their similarity to ancient Israelites and Jews rather than modern ones. Think about it the next time that your favorite genetic testing company tells you that you have some “Ashkenazic Jewish ancestry” or worse, that you are 100% Jews.
I hope that future tests covering other regions of the world would be able to explain the remaining portion of the elusive Jewish ancestry. Will “Genetic Jewishness” evolve onto “Primeval Jewishness” where people define their Jewishness based on their similarity to ancient Israelites and Jews rather than modern ones. Only time will say.
Dr. Eran Elhaik is a population, medical, and evolutionary geneticist at the University of Sheffield, UK. EranelHaikLab.org. All the rights to the content of this article are reserved for the author.
References
- Atzmon, G.L. HaoI. Pe’er et al. 2010. Abraham’s children in the genome era: major Jewish diaspora populations comprise distinct genetic clusters with shared Middle Eastern ancestry. American Journal of Human Genetics. 86:850-859.
- Behar, D. M.M. F. HammerD. Garrigan et al. 2004. MtDNA evidence for a genetic bottleneck in the early history of the Ashkenazi Jewish population. European Journal of Human Genetics. 12:355-364.
- Costa, M. D.J. B. PereiraM. Pala et al. 2013. A substantial prehistoric European ancestry amongst Ashkenazi maternal lineages. Nature Communications. 4:2543.
- Das, R., P. Wexler, M. Pirooznia, and E. Elhaik. 2016. Localizing Ashkenazic Jews to primeval villages in the ancient Iranian lands of Ashkenaz. Genome Biology and Evolution. 8:1132–1149.
- Das, R., P. Wexler, M. Pirooznia, and E. Elhaik. 2017. The Origins of Ashkenaz, Ashkenazic Jews, and Yiddish. Frontiers in genetics. 8.
- Efron, J. M. 1994. Defenders of the Race. Yale University Press, New Haven, CT.
- Elhaik, E. 2016. In search of the jüdische Typus: a proposed benchmark to test the genetic basis of Jewishness challenges notions of “Jewish biomarkers”. Frontiers in genetics. 7.
- Falk, R. 2017. Zionism and the Biology of the Jews. Springer, Cham, Switzerland.
- Kirsh, N. 2003. Population genetics in Israel in the 1950s. The unconscious internalization of ideology. Isis. 94:631-655.
- Matheson, C. D., K. K. Vernon, A. Lahti, R. Fratpietro, M. Spigelman, S. Gibson, C. L. Greenblatt, and H. D. Donoghue. 2009. Molecular Exploration of the First-Century Tomb of the Shroud in Akeldama, Jerusalem. PLOS ONE. 4:e8319.
- Nielsen, R., J. M. Akey, M. Jakobsson, J. K. Pritchard, S. Tishkoff, and E. Willerslev. 2017. Tracing the peopling of the world through genomics. Nature. 541:302-310.
- Ostrer, H. 2012. Legacy: a genetic history of the Jewish people. Oxford University Press, Oxford.
- Prohaska, A.F. RacimoA. J. Schork et al. 2019. Human Disease Variation in the Light of Population Genomics. Cell. 177:115-131.
- Skorecki, K., S. Selig, S. Blazer, R. Bradman, N. Bradman, P. J. Waburton, M. Ismajlowicz, and M. F. Hammer. 1997. Y chromosomes of Jewish priests. Nature. 385:32.

The question of Jewish ancestry has been the subject of controversy for over two centuries and has yet to be resolved. The “Rhineland Hypothesis” depicts Eastern European Jews as a “population isolate” that emerged from a small group of German Jews who migrated eastward and expanded rapidly.
Alternatively, the “Khazarian Hypothesis” suggests that Eastern European Jew descended from the Khazars, an amalgam of Turkic clans that settled the Caucasus in the early centuries CE and converted to Judaism in the 8th century.
Mesopotamian and Greco-RomanJews continuously reinforced the Judaized Empire until the 13 th century. Following the collapse of their empire, the Judeo-Khazars fled to Eastern Europe. The rise of European Jewry is therefore explained by the contribution of the Judeo-Khazars.
Thus far, however, the Khazar’scontribution has been estimated only empirically, as the absence of genome-wide data from Caucasus populations precluded testing the Khazarian Hypothesis. Recent sequencing of modern Caucasus populations prompted us to revisit the Khazarian Hypothesis and compare it with the Rhineland Hypothesis.
We applied a wide range of population genetic analyses to compare these two hypotheses. Our findings support the Khazarian Hypothesis and portray the European Jewish genome as a mosaic of Caucasus, European, and Semitic ancestries, thereby consolidating previous contradictory reports of Jewish ancestry.
We further describe the major differences among Caucasus populations explained by the early presence of Judeans in the Southern and CentralCaucasus. Our results have important implications on the demographic forces that shaped the genetic diversity in the Caucasus and medical studies.
Introduction
Contemporary Eastern European Jews comprise the largest ethnoreligious aggregate of modern Jewish communities, accounting for nearly 90% of over 13 million Jews worldwide (UnitedJewish Communities 2003).
Speculated to have emerged from a small Central European founder group and thought to have maintained high endogamy, Eastern European Jews are considered a“population isolate” and invaluable subjects in disease studies (Carmeli 2004), although their ancestry remains debatable between geneticists, historians, and linguists (Wexler 1993; Brook2006; Sand 2009; Behar et al. 2010).
Recently, several large-scale studies have attempted to chart the genetic diversity of Jewish populations by genotyping Eurasian Jewish and non-Jewish populations (Conrad et al. 2006; Kopelman et al. 2009; Behar et al. 2010). Interestingly, some of these studies linked Caucasus populations with Eastern European Jews, at odds with the narrative of a Central European founder group.
Because correcting for population structure and using suitable controls are critical in medical studies, it is vital to examine the hypotheses pertaining to explain the ancestry of Eastern and Central European Jews. One of the major challenges for any hypothesis is to explain the massive presence of Jews in Eastern Europe, estimated at eight million people at the beginning of the 20th century.
We investigate the genetic structure of European Jews, by applying a wide range of analyses — including 3 population test, principal component, biogeographical origin, admixture, identity by descent, allele sharing distance, and uniparental analyses — and test their veracity in light of the two dominant hypotheses depicting either a sole Middle Eastern ancestry or a mixed Middle Eastern-Caucasus-European ancestry to explain the ancestry of Eastern European Jews.
The “Rhineland Hypothesis” envisions modern European Jews to be the descendants of the Judeans – an assortment of Israelite-Canaanite tribes of Semitic origin (Figures 1,2)(Supplementary Note 1). It proposes two mass migratory waves: the first occurred over the two hundred years following the Muslim conquest of Palestine (638 CE) and consisted of devotedJudeans who left Muslim Palestine for Europe (Dinur 1961).
Whether these migrants joined the existing Judaized Greco-Roman communities is unclear, as is the extent of their contribution to 4the Southern European gene pool. The second wave occurred at the beginning of the 15 century by a group of 50,000 German Jews who migrated eastward and ushered an apparent hyper-baby- boom era for half a millennia (Atzmon et al. 2010).
The Rhineland Hypothesis predicts a MiddleEastern ancestry to European Jews and high genetic similarity among European Jews (Ostrer2001; Atzmon et al. 2010; Behar et al. 2010).

The competing “Khazarian Hypothesis” considers Eastern European Jews to be the descendants of Khazars (Supplementary Note 1).
The Khazars were a confederation of Slavic, Scythian, Hunnic-Bulgar, Iranian, Alans, and Turkish tribes who formed in the central-northern Caucasusone of the most powerful empires during the late Iron Age and converted to Judaism in the 8 century CE (Figures 1-2) (Polak 1951; Brook 2006; Sand 2009).
The Khazarian, Armenian, and Georgian populations forged from this amalgamation of tribes (Polak 1951) followed by relative isolation, differentiation, and genetic drift in situ (Balanovsky et al. 2011).
Biblical and archeological records allude to active trade relationships between Proto-Judeans and Armeniansin the late centuries BCE (Polak 1951; Finkelstein and Silberman 2002), which likely resulted in a small scale admixture between these populations and a Judean presence in the Caucasus.
After their conversion to Judaism, the population structure of the Judeo-Khazars was further reshaped by multiple migrations of Jews from the Byzantine Empire and Caliphate to the KhazarianEmpire (Figure 1). Following the collapse of their Empire and the Black Death (1347-1348) the Judeo-Khazars fled eastwards (Baron 1993), settling in the rising Polish Kingdom and Hungary(Polak 1951) and eventually spreading to Central and Western Europe.
The KhazarianHypothesis posits that European Jews are comprised of Caucasus, European, and Middle Eastern ancestries. Moreover, European Jewish communities are expected to be different from one another both in ancestry and genetic heterogeneity. The Khazarian Hypothesis also offers two explanations for the genetic diversity in Caucasus groups first by the multiple migration waves to Khazaria during the 6th to 10th centuries and second by the Judeo-Khazars who remained in the Caucasus.
5Genetic studies attempting to infer the ancestry of European Jews yielded inconsistent results. Some studies pointed to the genetic similarity between European Jews and the Caucasus populations like Adygei (Behar et al. 2003; Levy-Coffman 2005; Kopelman et al. 2009), while some pointed to the similarity to Middle Eastern populations such as Palestinians (Hammer et al. 2000; Nebelet al. 2000), and others pointed to the similarity to Southern European populations like Italians(Atzmon et al. 2010; Zoossmann-Diskin 2010).
Most of these studies were done in the pre-genomewide era using uniparental markers and included different reference populations, which makes it difficult to compare their results. More recent studies employing whole-genome data reported high genetic similarity of European Jews to Druze, Italian, and Middle Eastern populations (Atzmon et al. 2010; Behar et al. 2010).
Although both the Rhineland and Khazarian Hypotheses depict a Judean ancestry and are not mutually exclusive, they are well distinguished, as Caucasus and Semitic populations are considered ethnically and linguistically distinct (Patai and Patai 1975; Wexler 1993; Balanovskyet al. 2011).
Jews, according to either hypothesis, are an assortment of tribes who accepted Judaism, migrated elsewhere, and maintained their religion up to this date and are, therefore, expected to exhibit certain differences from their neighboring populations.
Because, according to both hypotheses, Eastern European Jews arrived in Eastern Europe roughly at the same time (13th and 15th centuries), we assumed that they experienced similar low and fixed admixture rates with the neighboring populations, estimated at 0.5% per generation over the past 50 generations(Ostrer 2001).
These relatively recent admixtures have likely reshaped the population structure of all European Jews and increased the genetic distances from the Caucasus or Middle Eastern populations. Therefore, we do not expect to achieve perfect matching with the surrogateKhazarian and Judean populations but rather to estimate their relatedness.
Materials and MethodsData collection
The complete data set contained 1,287 unrelated individuals of 8 Jewish and 74non-Jewish populations genotyped over 531,315 autosomal single nucleotide polymorphisms(SNPs). An LD-pruned data set was created by removing one member of any pair of SNPs in strong LD (r2 >0.4) in windows of 200 SNPs (sliding the window by 25 SNPs at a time) using indep-pairwise in PLINK (Purcell et al. 2007).
This yielded a total of 221,558 autosomal SNPs that were chosen for all autosomal analyses except the identical by descent analysis that utilized the complete data set. Both data sets were obtained fromhttp://www.evolution.ut.ee/MAIT/jew_data/ (Behar et al. 2010). MtDNA and Y-chromosomal data were obtained from previously published data sets as appear in Behar et al. (2010).
These markers were chosen to match the phylogenetic level of resolution achieved in previously reported data sets and represent a diversified set of markers. A total of 11,392 samples were assembled for mtDNA (6,089) and Y-chromosomal (5,303) analyses from 27 populations(Tables S1-2).
Terminology
In common parlance, Eastern and Central European Jews are practically synonymous with Ashkenazi Jews and are considered a single entity (Tian et al. 2008; Atzmon et al. 2010; Behar et al. 2010). However, the term is misleading, for the Hebrew word “Ashkenaz” was applied to Germany in medieval rabbinical literature – contributing to the narrative that modern Eastern European Jewry originated on the Rhine.
We thus refrained from using the term“Ashkenazi Jews.” Jews were roughly subdivided into Eastern (Belorussia, Latvia, Poland, and Romania) and Central (Germany, Netherland, and Austria) European Jews. In congruence with the literature that considers “Ashkenazi Jews” distinct from “Sephardic Jews,” we excluded the latter. Complete population notation is described in Table S3.
Choice of surrogate populations
As the ancient Judeans and Khazars have been vanquished and their remains have yet to be sequenced, in accordance with previous studies (Levy-Coffman 72005; Kopelman et al. 2009; Atzmon et al. 2010; Behar et al. 2010), contemporary MiddleEastern and Caucasus populations were used as surrogates.
Palestinians were considered proto-Judeans because they are assumed to share a similar linguistic, ethnic, and geographic background with the Judeans and were shown to share common ancestry with European Jews(Bonné-Tamir and Adam 1992; Nebel et al. 2000; Atzmon et al. 2010; Behar et al. 2010).
Similarly, Caucasus Georgians and Armenians were considered proto-Khazars because they are believed to have emerged from the same genetic cohort as the Khazars (Polak 1951; Dvornik1962; Brook 2006).
The 3 population test
The f3 statistics uses allele frequency differences to assess the presence of admixture in a population X from two other populations A and B, so that f3 (X; A, B)(Reich et al. 2009). If X is a mixture of A and B, rather than the result of genetic drift, f3 would be negative.
A significant negative f3 indicates that the ancestors of group X experienced a history of admixture subsequent to their divergence from A and B. The f3 statistics were calculated with the three pop programs ofTreeMix (Pickrell and Pritchard 2012) with k=500 over the set of 221,558 SNPs.
This test differs from ADXMITURE (Alexander, Novembre, and Lange2009), which reports the proportions of admixture with the most likely ancestor.

Principal component analysis (PCA)
Although the commonly used “multi-population” PCA has many attractive properties, it should be practiced with caution to avoid biases due to the choice of populations and varying sample sizes (Price et al. 2006; McVean 2009).
To circumvent these biases, we developed a simple “dual-population” framework consisting of three “outgroup” populations that are available in large sample sizes and are the least admixed — Mbuti and BiakaPygmies (South Africa), French Basques (Europe), and Han Chinese (East Asia) — and two populations of interest, all of the equal sample sizes.
The cornerstone of this framework is that it minimizes the number of significant PCs to four or fewer (Tracy-Widom test, p <0.01) and maximizes the portion of explained variance to over 20% for the first two PCs. PCA calculations were carried out using the smart PCA of the EIGENSOFT package (Patterson, Price, and Reich 2006).
Convex hulls were calculated using Matlab “convhull” function and plotted around the cluster centroids. Relatedness between two populations of interest was estimated by the commensurate overlap of their clusters. Small populations (<7 samples) were excluded from the analysis.
Estimating the biogeographical origins of the population
Novembre et al. (2008) proposed a PCA-based approach, accurate to a few hundred kilometers within Europe, to identify the current biogeographical origin of a population. Although this approach has no implied historical model, it correlates genetic diversity with geography and can thus be a useful tool to study biogeography.
To decrease the bias caused by multiple populations of uneven sizes (Patterson, Price, and Reich 2006; McVean 2009), we adopted the dual-population framework with three outgroup populations and two populations of interest: a population of known geographical origin during the relevant time period shown to cluster with the population in question (e.g., Armenians) and the population in question (e.g., Eastern European Jews).
The first four populations were used as a training set for the population in question. PCA calculations were carried out as described above. The rotation angle of PC1-PC2 coordinates was calculated as described by Novembre et al. (2008).
Briefly, in each figure, the PC axes were rotated to find the angle that maximizes the summed correlation of the median PC1 and PC2 values of the training populations with the latitude and longitude of their countries. Latitudinal and longitudinal data were obtained from the literature or by the country’s approximate centroid. Geodesic distances were calculated in kilometers using the Matlab function “distance.”
Admixture analysis
A structure-like approach was applied in a supervised learning model as implemented in;
- ADMIXTURE (Alexander, Novembre, and Lange 2009).
- ADMIXTURE provides an estimation of the individual’s ancestries from the allele frequencies of the designated ancestral populations.
- ADMIXTURE’s bootstrapping procedure with default parameters was used to calculate the standard errors. We observed low (<0.05) standard errors in all our analyses.
Populations were sorted by their mean African and Asian ancestries. In this analysis, the three Netherland Jews were grouped with Eastern European Jews.
Identity by descent (IBD) analysis.
To detect IBD segments, we ran fast IBD ten times using different random seeds and combined the results as described by Browning and Browning(2011). Segments were considered to be IBD only if the fast IBDscore of the combined analysis was less than e –10
This low threshold corresponds to long shared haplotypes (≥1 cM) that are likely to be IBD. Short gaps (<50 indexes) separating long domains were assumed to be false-negative and concatenated (Browning and Browning 2011). Pairwise-IBD segments between European Jews and different populations were obtained by finding the maximum total IBDsharing between each European Jew and all other individuals of a particular population.
Allele sharing distances (ASD)
ASD was used for measuring genetic distances between populations as it is less sensitive to small sample sizes than other methods. Pairwise ASD was calculated using PLINK (Purcell et al. 2007), and the average ASD between populations I and J, was computed as: I I J iij IJ nmW W/)(,(1)where W ij is the distance between individuals i and j from populations I and J of sizes n and m, respectively.
To verify that these ASD differences are significant, a bootstrap approach was used with the null hypothesis: H0: ASD ( p1, p2) = ASD ( p1, p3), where the ASD between populations p1 and p2 is compared to the ASD between populations p2 and p3(Supplementary Note 2). To compare continental Jewish communities, individuals were grouped by their continent and the comparison was carried as described.
Uniparental analysis.
To infer the migration patterns of European Jews, we integrated haplogroup data from over 11,300 uniparental chromosomes with geographical data. The haplogroup frequencies were compared between populations to obtain a measure of distance between populations.
Pairwise genetic distances between population haplogroups (Tables S1-2) were estimated by applying the Kronecker function as implemented in Arlequin version 3.1(Excoffier, Laval, and Schneider 2005).
In brief, the similarity between populations has defined as the fraction of haplogroups that the two populations shared as measured by the Kroneckerfunction)(i xy: Li xy xy id 1)(, (2)which equals 1 if the haplogroup frequency of the i-th haplogroup is non-zero for both populations and equals 0 otherwise.
In other words, populations sharing the same exact haplogroups or their mutual absence are considered more genetically similar than populations with different haplogroups. For brevity, we considered only haplogroups with frequencies higher than 0.5%.
This measure has several desirable properties that make it an excellent measure for estimating the genetic distance between populations, such as a simple interpretation in terms of homogeneity and applicability to both mtDNA and Y-chromosomal data.
Results
To confirm that the Rhineland and Khazarian Hypotheses indeed portray distinct ancestries, we assessed the degree of background admixture between Caucasus and Semitic populations. We calculated the f3 statistics between Palestinians and six of the Caucasus and Eurasian populations using African San as an outgroup, for example, f3 ( Palestinians, Armenians ).
The f3 results for Turks(–0.0013), Armenians and Georgians (–0.0019), Lezgins and Adygei (–0.0015), and Russians (– 0.0011) indicated a minor but significant admixture (–26<Z-score <–13) between Palestinians and the populations tested.
Because Armenians and Georgians diverged from Turks 600generations ago (Schonberg et al. 2011) we can assume that the lion’s share of their admixture derived from that ancestry and within the expected levels of background admixture typical to the region rather than recent admixture with Semitic populations. Therefore, similarities between European Jews and Caucasus populations will unlikely be due to shared Semitic ancestry.
PCA was next used to identify independent dimensions that capture most of the information in the data. PCA was applied using two frameworks: the “multi-population” carried for all populations (Figure 3) and separately for Eurasian populations along with Pygmies and Han Chinese (Figure S2) and our novel “dual-population” framework (Figure S3).
In all analyses, the studied samples aligned along the two well-established geographic axes of global genetic variation: PC1 (sub-Saharan Africa versus the rest of the Old World) and PC2 (east versus west Eurasia) (Li et al. 2008).
Our results reveal geographically refined groupings, such as the nearly symmetrical continuous European rim extending from Western to Eastern Europeans, the parallelCaucasus rim, and the Near Eastern populations (Figure S1) organized in Turk-Iranian and Druzeclusters (Figure 3). Middle Eastern populations form a gradient along the diagonal line between bedouin and Near Eastern populations that resemble their geographical distribution.
The remaining Egyptians and the bulk of Saudis distribute separately from Middle Eastern populations. European Jews are expected to cluster with native Middle Eastern or Caucasus populations according to the Rhineland or Khazarian Hypotheses, respectively. The results of all PC analyses(Figures 3, S2-3) show that over 70% of European Jews and almost all Eastern European Jewscluster with Georgian, Armenian, and Azerbaijani Jews within the Caucasus rim (Figures 3, S3).
Nearly 15% of Central European Jews cluster with Druze and the rest cluster with Cypriots. European Jews cluster distinctly from the Middle Eastern cluster. Strong evidence for the Khazarian Hypothesis is the clustering of European Jews with the populations that reside on opposite ends of ancient Khazaria: Armenians, Georgians, and Azerbaijani Jews (Figure 1).
Because Caucasus populations remained relatively isolated in the Caucasus region and because there are no records of Caucasus populations mass-migrating to Eastern and Central Europe prior to the fall of Khazaria (Balanovsky et al. 2011), these findings imply a shared origin for European Jews and Caucasus populations.
To assess the ability of our PCA-based approach to identify the biogeographical origins of a population, we first sought to identify the biogeographical origin of Druze. The Druze religion originated in the 11th century, but the people’s origins remain a source of much confusion and debate (Hitti 1928). We traced the Druze biogeographical origin to the geographical coordinates:38.6±3.45° N, 36.25±1.41° E (Figure S4) in the Near East (Figure S1).
Half of the Druzeclustered tightly in Southeast Turkey, and the remaining was scattered along with northern Syria and Iraq. These results are in agreement with Shlush et al.’s (2008) findings using mt DNA analysis. The inferred geographical positions of Druze were used in the subsequent analyses.
The geographical origins of European Jews varied for different reference populations (Figure 4, S5), but all the results converged to Southern Khazaria along with modern Turkey, Armenia, Georgia, and Azerbaijan. Eastern European Jews clustered tightly compared to Central European Jews in all analyses.
The smallest deviations in the geographical coordinates were obtained with Armenians for both Eastern (38±2.7° N, 39.9±0.4° E) and Central (35±5° N, 39.7±1.1° E)European Jews (Figure 4). Similar results were obtained for Georgians (Figure S5). Remarkably, the mean coordinates of Eastern European Jews are 560 kilometers from Khazaria’s southern border (42.77° N, 42.56° E) near Samandar – the capital city of Khazaria from 720 to 750 CE(Polak 1951).
The duration, direction, and rate of gene flow between populations determine the proportion of admixture and the total length of chromosomal segments that are identical by descent (IBD). Admixture calculations were carried out using a supervised learning approach in a structure-like analysis.
This approach has many advantages over the unsupervised approach that not only traces ancestry to K abstract unmixed populations under the assumption that they evolved independently (Chakravarti 2009; Weiss and Long 2009) but also is problematic when applied to study Jewish ancestry, which can be dated only as far back as 3,000 years (Figure 2).
Moreover, the results of the unsupervised approach vary based on the particular populations used for the analysis and the choice of K, rendering the results incomparable between studies.
The admixture was calculated with a reference set of seven populations representing genetically distinct regions: Pygmies (South Africa), French Basque (West Europe), Chuvash (East Europe), Han Chinese(East Asia), Palestinians (Middle East), Turk-Iranians (Near East), and Armenians (Caucasus)(Figure 5).
The ancestral components grouped all populations by their geographical regions with European Jews clustering with Caucasus populations. As expected, Eastern and WesternEuropean ancestries exhibit opposite gradients among European populations.
The Near Eastern-Caucasus ancestries are dominant among Central (38%) and Eastern (32%) European Jews followed by Western European ancestry (30%). Among non-Caucasus populations, the Caucasusancestry is the largest among European Jews (26%) and Cypriots (31%). These populations also exhibit the largest fraction of Middle Eastern ancestry among non-Middle Eastern populations.
As both Caucasus and Middle Eastern ancestries are absent in Eastern European populations, our findings suggest that Eastern European Jews acquired these ancestries prior to their arrival in eastern Europe.
Although the Rhineland Hypothesis explains the Middle Eastern ancestry by stating that Jews migrated from Palestine to Europe in the 7th century, it fails to explain the large Caucasus ancestry, which is nearly endemic to Caucasus populations.
Although they cluster with Caucasus populations (Figure 5), Eastern and Central European Jews share a large fraction of Western European and Middle Eastern ancestries, both absent in Caucasus populations, excepting Armenians who share 15±2% Middle Eastern ancestry (FigureS6).
According to the Khazarian Hypothesis, the Western European ancestry was imported to Khazaria by Greco-Roman Jews, whereas the Middle Eastern ancestry alludes to the contribution of both early Israelite Proto-Judeans as well as Mesopotamian Jews (Polak 1951; Koestler 1976; Sand 2009).
Central and Eastern European Jews differ mostly in their MiddleEastern (30% and 25%, respectively) and Eastern European ancestries (3% and 12%, respectively), probably due to late admixture.
Druze exhibits a large Turkic ancestry (83%) in accordance with their Near Eastern origin (FigureS4). Druze and Cypriot appear similar to European Jews in their Middle Eastern and Western European ancestries, though they differ largely in the proportion of Caucasus ancestry.
These results can explain the genetic similarity between European Jews, Southern Europeans, and Druze reported in studies that excluded Caucasus populations (Price et al. 2008; Atzmon et al.2010; Zoossmann-Diskin 2010). Overall, our results portray the European Jewish genome as a mosaic of the Caucasus, Western European, Middle Eastern, and Eastern European ancestries in decreasing proportions.
To glean further details of the genomic regions contributing to the genetic similarity between European Jews and the perspective populations, we compared their total genomic regions shared by IBD.
If European Jews emerged from Caucasus populations, the two would share longer IBDregions than with Middle Eastern populations. The IBD analysis exhibits a skewed bimodal distribution embodying a major Caucasus ancestry with a minor Middle Eastern ancestry (Figure6), consistent with the admixture results (Figure 5).
he total IBD regions shared between European Jews and Caucasus populations (9.5 cm on average) are significantly larger than regions shared with Palestinians (5.5 cM) (Kolmogorov-Smirnov goodness-of-fit test, p <0.001).
To the best of our knowledge, these are the largest IBD regions ever reported between EuropeanJews and non-Jewish populations. The decrease in total IBD between European Jews and other populations combined with the increase in distance from the Caucasus supports the KhazarianHypothesis.

We next estimated the level of endogamy among Eurasian Jewish communities and compared their genetic distances to non-Jewish neighbors, Caucasus, and Middle Eastern populations. Our results expand the previous report of high endogamy in Jewish populations (Behar et al. 2010)and narrow the endogamy to regional Jewish communities (Table 1, left panel).
Jews are significantly more similar to members of their own community than to other Jewish populations(P<0.01, bootstrap -test), with the conspicuous exception of Bulgarian, Turkish, and GeorgianJews. These results stress the high heterogeneity among Jewish communities across Eurasia and even within communities, as in the case of the Balkan and Caucasus Jews.
When compared to non-Jewish populations, all Jewish communities were significantly (P <0.01, bootstrap t-test) distant from Middle Eastern populations and, with the exception of central European Jews, significantly closer to Caucasus populations (Table 1, right panel). Similar findings were reported by Behar et al. (2010) although they were dismissed as “a bias inherent in our calculations.“
The close genetic distance between Central European Jews and SouthernEuropean populations can be attributed to a late admixture. The results are consistent with our previous findings in support of the Khazarian Hypothesis.
As the only commonality among Jewish communities is their dissimilarity from Middle Eastern populations (Table 1), grouping different Jewish communities without correcting for their country of origin, as is commonly done, would increase their genetic heterogeneity. Finally, we carried out uniparental analyses on mtDNA and Y-chromosome comparing the haplogroup frequencies between European Jews and other populations.
The Rhineland Hypothesis depicts Middle Eastern origins for European Jews’ both paternal and maternal ancestries, whereas the Khazarian Hypothesis depicts a Caucasus ancestry along with SouthernEuropean and Near Eastern contributions of migrates from Byzantium and the Caliphate, respectively.
Because Judaism was maternally inherited only since the 3rd century CE (Patai and Patai 1975), the mtDNA is expected to show a stronger local female-biased founder effect compared to the Y-chromosome. Haplogroup similarities between European Jews and other populations were plotted as heat maps on the background of their geographical locations (Figure7). The pairwise distances between all studied populations are shown in Figure S7.
Our results shed light on sex-specific processes that, although not evident from the autosomal data, are analogous to those obtained from the biparental analyses. Both mtDNA and Y-chromosomal analyses yield high similarities between European Jews and Caucasus populations rooted in the Caucasus (Figure 7) in support of the Khazarian Hypothesis.
Interestingly, the maternal analysis depicts a specific Caucasus founding lineage with a weak Southern European ancestry (Figure 7a), whereas the paternal ancestry, reveals a dual Caucasus-Southern European origin (Figure 7b). As expected, the maternal ancestry exhibits a higher relatedness scale with narrow dispersal compared with the paternal ancestry. Dissecting uniparental haplogroups allows us to delve further into European Jews’ migration routes.
As the results do not specify whether the Southern Europe-Caucasus migration was ancient or recent nor indicate the migration’s direction i.e., from Southern Europe to the Caucasus or the opposite, there are four possible scenarios.
Of these, the only historically supportable scenarios are ancient migrations from Southern Europe toward Khazaria (6th-13th centuries) and more recent migrations from the Caucasus to Central and Southern Europe (13th-15th centuries) (Polak 1951; Patai and Patai 1975; Straten 2003; Brook 2006; Sand 2009).
A westward migration from the diminished Khazaria toward Central and Southern Europe would have exhibited a gradient from the Caucasus toward Europe for both matrilineal and patrilineal lines. Such a gradient was not observed.
By contrast, Judaized Greco-Roman male-driven migration directly to Khazaria is consistent with historical demographic migrations and could have created the observed pattern. Moreover, we found little genetic similarity between European Jews and populations eastward to the Caspian Sea and southward to the Black Sea, delineating the geographical boundaries of Khazaria (Table 1, Figure 1).

Discussion
Eastern and Central European Jews comprise the largest group of contemporary Jews, accounting for nearly 90% of over 13 million worldwide Jews (United Jewish Communities2003). Eastern European Jews made up over 90% of European Jews before World War II.
Despite their controversial ancestry, European Jews are an attractive group for genetic and medical studies due to their presumed genetic history (Ostrer 2001). Correcting for population structure and using suitable controls are critical in medical studies, thus it is vital to determine whether European Jews are of Semitic, Caucasus, or another ancestry.
Though Judaism was born encased in theological-historical myth, no Jewish historiography was produced from the time of Josephus Flavius (1st century CE) to the 19th century (Sand 2009). Early historians bridged the historical gap simply by linking modern Jews directly to the ancient Judeans (Figure 2); a paradigm that was later embedded in medical science and crystallized as narrative.
Many have challenged this narrative (e.g., Koestler 1976; Straten 2007), mainly by showing that a sole Judean ancestry cannot account for the vast population of Eastern EuropeanJews at the beginning of the 20th century without the major contribution of Judaized Khazars and by demonstrating that it is in conflict with anthropological, historical, and genetic evidence(Patai and Patai 1975; Baron 1993; Sand 2009).
With uniparental and whole-genome analyses providing ambiguous answers (Levy-Coffman2005; Atzmon et al. 2010; Behar et al. 2010), the question of European Jewish ancestry remained debated between the supporters of the Rhineland and Khazarian Hypotheses.
Although both theories oversimplify complex historical processes they are attractive due to their distinct predictions and testable hypotheses. We showed that the hypotheses are also genetically distinct and that the minuscule Semitic ancestry in Caucasus populations cannot account for the similarity between European Jews and Caucasus populations.
The recent availability of genomic data from Caucasus populations allowed testing the Khazarian Hypothesis for the first time and prompted us to contrast it with the Rhineland Hypothesis.
To evaluate the two hypotheses, we carried out a series of comparative analyses between European Jews and surrogate Khazarian and Judean populations posing the same question each time: are Eastern and Central European Jewsgenetically closer to Khazarian or Judean populations?
Under the Rhineland Hypothesis, European Jews are also expected to exhibit high endogamy across all their communities and be more similar to Middle Eastern populations compared to their neighboring non-Jewish populations, whereas the Khazarian Hypothesis predicts the opposite scenario. We emphasize that these hypotheses are not exclusive and that some European Jews may have other ancestries.
Our PC, biogeographical estimation, admixture, IBD, ASD, and uniparental analyses were consistent in depicting a Caucasus ancestry for European Jews. Our first analyses revealed a tight genetic relationship of European Jews and Caucasus populations and pinpointed the biogeographical origin of European Jews to the south of Khazaria (Figures 3,4).
Our later analyses yielded a complex ancestry with a slightly dominant Near Eastern-Caucasus ancestry, large Southern European and Middle Eastern ancestries, and a minor Eastern European contribution; the latter two differentiated Central and Eastern European Jews (Figures 4, 5 and Table 1).
While the Middle Eastern ancestry faded in the ASD and uniparental analyses, theSouthern European ancestry was upheld probably attesting to its later time period (Table 1 andFigure 7). We show that the Khazarian Hypothesis offers a comprehensive explanation for the results, including the reported Southern European (Atzmon et al. 2010; Zoossmann-Diskin 2010) and middle Eastern ancestries (Nebel et al. 2000; Behar et al. 2010).
By contrast, the RhinelandHypothesis could not explain the large Caucasus component in European Jews, which is rare in Non-Caucasus populations (Figure 5), and the large IBD regions shared between European Jews and Caucasus populations attesting to their common origins. Our findings thus reject the Rhineland Hypothesis and uphold the thesis that Eastern European Jews are Judeo-Khazars in origin.
Consequently, we can conclude that the conceptualization of European Jews as a“population isolate,” which is derived from the Rhineland Hypothesis, is incorrect and most likely reflects sampling bias in the lack of Caucasus non-Jewish populations in comparative analyses.
A major difficulty with the Rhineland Hypothesis, in addition to the lack of historical and anthropological evidence to the multi-migration waves from Palestine to Europe (Straten 2003; Sand 2009), is to explain the vast population expansion of Eastern European Jews from 50thousand (15th century) to 8 million (20th century).
The annual growth rate that accounts for this population expansion was estimated at 1.7-2%, one order of magnitude larger than that of eastern European non-Jews in the 15th-17th centuries, prior to the industrial revolution (Straten2007).
This growth could not possibly be the product of natural population expansion, particularly one subjected to severe economic restrictions, slavery, assimilation, the Black Death and other plagues, forced and voluntary conversions, persecutions, kidnappings, rapes, exiles, wars, massacres, and pogroms (Koestler 1976; Straten 2003; Sand 2009).
Because such a 19unnatural growth rate, over half a millennia and affecting only Jews residing in Eastern Europe,is implausible – it is explained by a miracle (Atzmon et al. 2010; Ostrer 2012). Unfortunately, this divine intervention explanation poses a new kind of problem – it is not science.
The question of how the Rhineland Hypothesis, so deeply rooted in supernatural reasoning, became the dominant scientific narrative is debated among scholars (e.g., Sand 2009). The most parsimonious explanation for our findings is that Eastern European Jews are of Judeo-Khazarian ancestry forged over many centuries in the Caucasus.
Jewish presence in the Caucasus and later Khazaria was recorded as early as the late centuries BCE and reinforced due to the increase in trade along the Silk Road (Figure 1), the decline of Judah (1st -7th centuries), and the uprise of Christianity and Islam (Polak 1951).
Greco-Roman and Mesopotamian Jews gravitating toward Khazaria were also common in the early centuries and their migrations were intensified following the Khazars’ conversion to Judaism (Polak 1951; Brook 2006; Sand 2009). The eastward male-driven migrations (Figure 7) from Europe to Khazaria solidified the exotic southern European ancestry in the Khazarian gene pool, (Figure 5) and increased the genetic heterogeneity of the Judeo-Khazars.
The religious conversion of the Khazars encompassed most of the Empire’s citizens and subordinate tribes and lasted for the next 400 years (Polak 1951; Baron 1993) until the invasion of the Mongols (Polak 1951; Dinur 1961; Brook 2006).
At the final collapse of their empire (13th century), many of the Judeo-Khazars fled to Eastern Europe and later migrated to Central Europe and admixed with the neighboring populations. Historical and archeological findings shed light on the demographic events following the Khazar’s conversion.
During the half-millennium of their existence (740-1250 CE), the Judeo-Khazars sent offshoots into the Slavic lands, such as Romania and Hungary (Baron 1993), planting the seeds of a great Jewish community to later rise in the Khazarian diaspora.
We hypothesize that the settlement of Judeo-Khazars in Eastern Europe was achieved by serial founding events, whereby populations expanded from the Caucasus into Eastern and Central Europe by successive splits, with daughter populations expanding to new territories following changes in socio-political conditions (Gilbert 1993). These events may have contributed to the higher homogeneity observed in Jewish communities outside Khazaria’s borders (Table 1).
After the decline of their Empire, the Judeo-Khazars refugees sought shelter in the emergingPolish Kingdom and other Eastern European communities where their expertise in economics, finances, and politics was valued. Prior to their exodus, the Judeo-Khazar population was estimated to be half a million in size, the same as the number of Jews in the Polish-Lithuaniankingdom four centuries later (Polak 1951; Koestler 1976).
Some Judeo-Khazars were left behind, mainly in the Crimea and the Caucasus, where they formed Jewish enclaves surviving into modern times. One of the dynasties of Jewish princes ruled in the 15th century under the tutelage of the Genovese Republic and later of the Crimean Tartars. Another vestige of the Khazar nation is the “Mountain Jews” in the North-Eastern Caucasus (Koestler 1976).
The remarkable close proximity of European Jews and populations residing on the opposite ends of ancient Khazaria, such as Armenians, Georgians, Azerbaijani Jews, and Druze (Figures 3, S2-3, 5), supports a common Near Eastern-Caucasus ancestry. These findings are not explained by the Rhineland Hypothesis and are staggering due to the uneven demographic processes these populations have experienced in the past eight centuries.
The high genetic similarity between European Jews and Armenian compared to Georgians (Figures 5 and 6, Table 1) is particularly bewildering because Armenians and Georgians are very similar populations that share a similar genetic background (Schonberg et al. 2011) and long history of cultural relations (Payaslian2007).
We identified a small Middle Eastern ancestry in Armenians that does not exist with Georgians and is likely responsible for the high genetic similarity between Armenians and European Jews (Figure S6).
Because the Khazars blocked the Arab approach to the Caucasus, we suspect that this ancestry was introduced by the Judeans arriving at a very early date to Armenia and was absorbed into the populations, whereas Judeans arriving in Georgia avoided assimilation(Shapira 2007). Similarly, the relatedness between European Jews and Druze reported here and in the literature (Behar et al. 2010) is explained by Druze Turkish-Southern Caucasus origins.
Druze migrated to Syria, Lebanon, and eventually to Palestine between the 11th and 13th centuries during the Crusades, a time when the Jewish population in Palestine was at a minimum. The genetic similarity between European Jews and Druze, therefore, supports the KhazarianHypothesis and should not be confused with a Semitic origin, which can be easily distinguished from the non-Semitic origin (Figure 5).
We emphasize that testing the Middle Eastern origin of European Jews can only be done with indigenous Middle Eastern groups. Overall, the similarity between European Jews and Caucasus populations underscores the genetic continuity that exists among Eurasian Jewish and non-Jewish Caucasus populations.
This genetic continuity is not surprising. The Caucasus gene pool proliferated from the NearEastern pool due to an Upper Paleolithic (or Neolithic) migration and was shaped by significant genetic drift, due to relative isolation in the extremely mountainous landscape (Balanovsky et al.2011; Pagani et al. 2011).
Caucasus populations are therefore expected to be genetically distinct from Southern European and Middle Eastern populations (Figure 5) but to share certain genetic similarities with Near Eastern populations such as Turks, Iranians, and Druze. In all our analyses, Middle Eastern samples clustered together or exhibited high similarity along a geographical gradient (Figure 3) and were distinguished from Arabian Peninsula Arab samples on one hand and from Near Eastern – Caucasus samples on the other hand.
Our study attempts to shed light on the forgotten Khazars and elucidate some of the most fascinating questions of their history. Although the Khazars’ conversion to Judaism is not in dispute, there are questions as to how widespread and established the new religion became.
Despite the limited sample size of European Jews, they represent members from the major residential Jewish countries (i.e., Poland and Germany) and exhibit very similar trends. Our findings support a large-scale migration from South-Central Europe and Mesopotamia to Khazaria that reshaped the genetic structure of the Khazars and other Caucasus populations in the central and upper Caucasus.
Another intriguing question touches upon the origins of the Khazars, speculated to be Turk, Tartar, or Mongol (Brook 2006). As expected from their common origin, Caucasus populations exhibit high genetic similarity to Iranian and Turks with mild Eastern Asian ancestry (Figure 5, Figure S7).
However, we found a weak patrilineal Turkiccontribution compared to the Caucasus and Eastern European contributions (Figure 7). Our findings thus support the identification of Turks as the Caucasus’ ancestors but not necessarily the predominant ancestors. Given their geographical position, it is likely the Khazarian gene pool was also influenced by Eastern European populations that are not represented in our dataset.
Our results fit with evidence from a wide range of fields. Linguistic findings depict EasternEuropean Jews as descended from a minority of Israelite-Palestinian Jewish emigrates who intermarried with a larger heterogeneous population of converts to Judaism from the Caucasus, the Balkans, and the Germano-Sorb lands (Wexler 1993).
Yiddish, the language of Central and Eastern European Jews, began as a Slavic language that was respecified to High German at an early date (Wexler 1993). Our findings are also in agreement with archeological, historical, linguistic, and anthropological studies (Polak 1951; Patai and Patai 1975; Wexler 1993; Brook2006; Kopelman et al. 2009; Sand 2009) and reconcile contradicting genetic findings observed uniparental and biparental genome data.
The conclusions of the latest genome-wide studies(Atzmon et al. 2010; Behar et al. 2010) that European Jews had a single Middle Eastern originate incomplete as neither study tested the Khazarian Hypothesis, to the extent done here.
Finally, our findings confirm both oral narratives and the canonical Jewish literature describing the Khazar’s conversion to Judaism (e.g., “Sefer ha-Kabbalah” by Abraham ben Daud [1161 CE], and “The Khazars” by Rabbi Jehudah Halevi [1140 CE]) (Polak 1951; Koestler 1976).
Although medical studies were not conducted using Caucasus and Near Eastern populations to the same extent as with European Jews, many diseases found in European Jews are also found in their ancestral groups in the Caucasus (e.g., Cystic fibrosis and α-thalassemia), the Near East(e.g., Factor XI deficiency, type II), and Southern Europe (e.g., Non-syndromic recessive deafness) (Ostrer 2001), attesting to their complex multi-origins.
Because our study is the first to directly contrast the Rhineland and Khazarian Hypotheses, caution is warranted in interpreting some of our results due to small sample sizes and availability of surrogate populations. To test the Khazarian Hypothesis, we used a crude model for the Khazar’s population structure. Our admixture analysis suggests that certain ancestral elements in the Caucasus genetic pool may have been unique to the Khazars.
Therefore, using few contemporary Caucasus populations as surrogates may capture only certain shades of the Khazarian genetic spectrum. Further studies are necessary to test the magnitude of the Judeo-Khazar demographic contribution to the presence of Jews in Europe (Polak 1951; Dinur 1961; Koestler 1976; Baron 1993; Brook 2006). These studies may yield a more complex demographic
23model than the one tested here and illuminate the complex population structure of Caucasus populations. Irrespective of these limitations, our results were robust across diverse types of analyses, and we hope that they will provide new perspectives for genetic, disease, medical, and anthropological studies.

Conclusions
We compared two genetic models for European Jewish ancestry depicting mixed Khazarian-European-Middle Eastern and sole Middle Eastern origins. Contemporary populations were used as surrogates to the ancient Khazars and Judeans, and their relatedness to European Jews was compared over a comprehensive set of genetic analyses.
Our findings support the Khazarian hypothesis depicting a large Caucasus ancestry along with Southern European, Middle Eastern, and Eastern European ancestries, in agreement with recent studies and oral and written traditions.
We conclude that the genome of European Jews is a tapestry of ancient populations including Judaized Khazars, Greco-Romans Jews, Mesopotamian Jews, and Judeans and that their population structure was formed in the Caucasus and the banks of the Volga with roots stretching to Canaan and the banks of the Jordan.

Supplemental Materials
Supplementary Note 1Supplementary Note 2Supplementary figures table S1 – Mt HaplogroupsTable S2 – Y HaplogroupsTable S3 – Studied PopulationsTable S4 – ASD analysis for Non-JewsTable S5 – ASD analysis for JewsTable S6 – Population Summary of ASD analysis table S7 – ASD results
Acknowledgment
I am grateful to Brian and Sharon Browning for their help with the IBD analysis, to my colleagues for their valuable comments. I thank two anonymous reviewers for their constructive comments and suggestions.
Reference
Alexander DH, Novembre J, Lange K. 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19:1655-1664.Atzmon G, Hao L, Pe’er I, et al. [11 co-authors]. 2010. Abraham’s children in the genome era: major Jewish diaspora populations comprise distinct genetic clusters with shared MiddleEastern Ancestry. Am. J. Hum. Genet. 86:850-859.Balanovsky O, Dibirova K, Dybo A, et al. [24 co-authors]. 2011. Parallel Evolution of Genes and Languages in the Caucasus Region. Mol. Biol. Evol. 28:2905-2920.Baron SW. 1993.
A Social and Religious History of the Jews. New York: Columbia UniversityPress.Behar DM, Thomas MG, Skorecki K, et al. [12 co-authors]. 2003. Multiple origins of Ashkenazi Levites: Y chromosome evidence for both Near Eastern and European ancestries. Am. J.Hum. Genet. 73:768-779.Behar DM, Yunusbayev B, Metspalu M, et al. [21 co-authors]. 2010. The genome-wide structure of the Jewish people. Nature. 466:238-242.Bonné-Tamir B, Adam A. 1992. Genetic diversity among Jews: diseases and markers at the DNA level. New York: Oxford University Press.Brook KA. 2006.
The Jews of Khazaria. Lanham, MD: Rowman & Littlefield.Browning BL, Browning SR. 2011. A fast, powerful method for detecting identity by descent. Am. J. Hum. Genet. 88:173-182.Carmeli DB. 2004. Prevalence of Jews as subjects in genetic research: Figures, explanation, and potential implications. Am. J. Med. Genet. A. 130A:76-83.Chakravarti A. 2009. Being human: kinship: race relations. Nature. 457:380-381.Conrad DF, Jakobsson M, Coop G, Wen X, Wall JD, Rosenberg NA, Pritchard JK. 2006.
A worldwide survey of haplotype variation and linkage disequilibrium in the human genome. Nat. Genet. 38:1251-1260.Dinur BZ. 1961. Israel in Exile (in Hebrew). Tel Aviv, Israel: Devir and Mosad Bialik.Dvornik F. 1962. The Slavs in European history and civilization. New Brunswick, NJ: RutgersUniversity Press.Excoffier L, Laval G, Schneider S. 2005. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol. Bioinform. Online. 1:47-50.Finkelstein I, Silberman NA. 2002. The Bible unearthed: archaeology’s new vision of ancient Israel and the origin of its sacred texts. New York, NY: Simon and Schuster.Gilbert M. 1993.
The atlas of Jewish history. New York: William Morrow and Company.Hammer MF, Redd AJ, Wood ET, et al. [12 co-authors]. 2000. Jewish and Middle Eastern non-Jewish populations share a common pool of Y-chromosome biallelic haplotypes. Proc. Natl. Acad. Sci. USA. 97:6769-6774.Hitti PK. 1928. The origins of the Druze people and religion, with extracts from their sacred writings. New York: Columbia University Press.Koestler A. 1976. The thirteenth tribe: the Khazar empire and its heritage. New York: RandomHouse.Kopelman NM, Stone L, Wang C, Gefel D, Feldman MW, Hillel J, Rosenberg NA. 2009.Genomic microsatellites identify shared Jewish ancestry intermediate between MiddleEastern and European populations. BMC Genet. 10:80. 26Levy-Coffman E. 2005.
A mosaic of people: the Jewish story and a reassessment of the DNA evidence. J. Genetic Genealogy. 1:12-33.Li JZ, Absher DM, Tang H, et al. [11 co-authors]. 2008. Worldwide human relationships are inferred from genome-wide patterns of variation. Science. 319:1100-1104.McVean G. 2009.
A genealogical interpretation of principal components analysis. PLoS Genet.5:e1000686. Nebel A, Filon D, Weiss DA, Weale M, Faerman M, Oppenheim A, Thomas MG. 2000. High-resolution Y chromosome haplotypes of Israeli and Palestinian Arabs reveal geographic substructure and substantial overlap with haplotypes of Jews. Hum. Genet. 107:630-641. Novembre J, Johnson T, Bryc K, et al. [12 co-authors]. 2008.
Genes mirror geography within Europe. Nature. 456:98-101.Ostrer H. 2001. A genetic profile of contemporary Jewish populations. Nat. Rev. Genet. 2:891-898.Ostrer H. 2012. Legacy: A Genetic History of the Jewish People. OUP USA.Pagani L, Ayub Q, Macarthur DG, et al. [13 co-authors]. 2011. High altitude adaptation in Daghestani populations from the Caucasus. Hum. Genet.:1-11.Patai R, Patai J. 1975. The myth of the Jewish race. New York: Scribner.Patterson N, Price AL, Reich D. 2006.
Population structure and eigenanalysis. Pp. e190. PLoSGenet.Payaslian S. 2007. The history of Armenia: from the origins to the present. New York: PalgraveMacmillan.Pickrell JK, Pritchard JK. 2012. Inference of population splits and mixtures from genome-wide allele frequency data. arXiv. 1206.2332v1.Polak AN. 1951. Khazaria – The History of the Jewish Kingdom in Europe (in Hebrew). Tel-Aviv, Israel: Mosad Bialik and Massada Publishing Company.Price AL, Butler J, Patterson N, et al. [20 co-authors]. 2008.
Discerning the ancestry of European Americans in genetic association studies. PLoS Genet. 4:e236.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. 2006. Principal components analysis corrects for stratification in genome-wide association studies. Nat.Genet. 38:904-909.Purcell S, Neale B, Todd-Brown K, et al. [11 co-authors]. 2007. PLINK: a toolset for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81:559-575.Reich D, Thangaraj K, Patterson N, Price AL, Singh L. 2009.
Reconstructing Indian population history. Nature. 461:489-494.Sand S. 2009. The invention of the Jewish people. London: Verso.Schonberg A, Theunert C, Li M, Stoneking M, Nasidze I. 2011. High-throughput sequencing of complete human mtDNA genomes from the Caucasus and West Asia: high diversity and demographic inferences. Eur. J. Hum. Genet. 19:988-994.Shapira DDY. 2007. Armenian and Georgian sources on the Khazars: a re-evaluation
In: Golden,PB, Ben-Shammai, H., and Róna-Tas, A., editor. The World of the Khazars: NewPerspectives – Selected Papers from the Jerusalem 1999 International KhazarColloquium. Leiden, Boston: Brill. p. 291-306.
Shlush LI, Behar DM, Yudkovsky G, Templeton A, Hadid Y, Basis F, Hammer M, Itzkovitz S,Skorecki K. 2008. The Druze: a population genetic refugium of the Near East. PLoS One.3:e2105.Straten JV. 2007. Early Modern Polish Jewry The Rhineland Hypothesis Revisited. Hist.Methods. 40:39-50.Straten JV. 2003.
Jewish Migrations from Germany to Poland: the Rhineland HypothesisRevisited. Mankind Quarterly. 44:367-384.Tian C, Plenge RM, Ransom M, et al. [11 co-authors]. 2008. Analysis and application of European genetic substructure using 300 K SNP information. PLoS Genet. 4:e4.United Jewish Communities. 2003. National Jewish Population Survey, 2000-01. New York: United Jewish Communities.Weiss KM, Long JC. 2009.
Non-Darwinian estimation: my ancestors, my genes’ ancestors.Genome Res. 19:703-710.Wexler P. 1993. The Ashkenazic Jews: A Slavo-Turkic People in Search of a Jewish Identity.Colombus, OH: Slavica.Zoossmann-Diskin A. 2010. The origin of Eastern European Jews was revealed by autosomal, sex chromosomal, and mtDNA polymorphisms. Biol Direct. 5:57.

Tables
Table 1.
Genetic distances (ASD) between regional and continental Jewish communities(left panel) and between regional Jewish communities and their non-Jewish neighboring populations, Caucasus, and Middle Eastern populations (right panel). Bold entries are significantly smaller throughout each panel.
The geographically nearest non-Jewish population was considered a neighboring population. The distances in the last two columns are between a Jewish and one Caucasus (Armenians of Georgians) or Middle Eastern (Palestinians, Bedouins,or Jordanians) population that exhibited the lowest mean ASD.
0.2318
0.2328 0.2381 0.2446 Hungarian 0.2346
0.2340
0.2387Central European
0.2312
0.2326 0.2378 0.2445 Italians
0.2335
0.2338 0.2385Bulgarian 0.2326 0.2331 0.2376 0.2439 Romanian 0.2347
0.2337
0.2380Turkish 0.2336 0.2336 0.2376 0.2439 Turkish 0.2353
0.2337
0.2379Iraqi
0.2303
0.2351 0.2375 0.2447 Iranian 0.2363
0.2338
0.2381Georgian 0.2304 0.2345 0.2372 0.2442 Georgian 0.2332
0.2332
0.2378Azerbaijani
0.2304
0.2365 0.2386 0.2465 Lezgins 0.2367
0.2352
0.2398Iranian
0.2310
0.2364 0.2391 0.2434 Iranian 0.2414
0.2361
0.2383
Figure legends
Figure 1
Map of Eurasia. A map of Khazaria and Judah is shown with the state of origin of the studied groups. Eurasian Jewish and non-Jewish populations used in all analyses are shown in square and round bullets, respectively (see Table S3). The major migrations that formed EasternEuropean Jewry according to the Khazarian and Rhineland Hypotheses are shown in yellow and brown, respectively.
Figure 2
An illustrated timeline for the relevant historical events. The horizontal dashed lines represent controversial historical events explained by the different hypotheses, whereas solid black lines represent undisputed historical events.
Figure 3
Scatter plot of all populations along with the first two principal components. For brevity, we show only the populations relevant to this study. The inset magnifies Eurasian and MiddleEastern individuals. Each letter code corresponds to one individual (Table S3). A polygon surrounding all of the individual samples belonging to a group designation highlights several population groups.
Figure 4
Biogeographical origin of European Jews. First, two principal components were calculated for pygmies, French Basque, Han Chinese (black), Armenians (blue), and Eastern or CentralEuropean Jews (red) – all of the equal size. PCA was calculated separately for Eastern and Central. European Jews and the results were merged. Using the first four populations as a training set, Eastern (squares) and Central (circles) European Jews were assigned to geographical locations by fitting independent linear models for latitude and longitude as predicted by PC1 and PC2. Each shape represents an individual. Major cities are marked in cyan.
Figure 5
Admixture analysis of European, Caucasus, Near Eastern, and Middle Eastern populations. The x-axis represents individuals from populations sorted according to their ancestries and arrayed geographically roughly from North to South. Each individual is represented by a vertically stacked column (100%) of color-coded admixture proportions of the ancestral populations.
Figure 6
The proportion of total IBD sharing between European Jews and different populations. Populations are sorted by decreasing distance from the Caucasus. The maximal IBD between each European Jew and an individual from each population is summarized in box plots. Lines pass through the mean values.
Figure 7
Pairwise genetic distances between European Jews and other populations measured across) mtDNA and b) Y-chromosomal haplogroup frequencies. The values of 1-xy are color-coded in a heat map with darker colors indicating higher haplogroup similarity with EuropeanJews.



2013




ATTENTION READERS
We See The World From All Sides and Want YOU To Be Fully InformedIn fact, intentional disinformation is a disgraceful scourge in media today. So to assuage any possible errant incorrect information posted herein, we strongly encourage you to seek corroboration from other non-VT sources before forming an educated opinion.
About VT - Policies & Disclosures - Comment Policy




The ongoing squabble between Israel and Iran can accurately be called a sibling rivalry.
The Biblical Patriarchs originated in Haran and Ur (Sanli Urfa) north of Aleppo & the Euphrate,
i.e. in the Southern Caucasus. This is in sharp conrast zo the Amalekite aka Bedouin cited in the article
for a reference of origin. Thus the research actually confirms that Ashkenazi Jews areN’T Amalekite,
while the Arab Jews are Arab/Amalekite. Israeli Ashkenazi Jews have lots of Armenian-like (south Caucasian) Haplogroups, e.g. Bibi, much more than American Ashkenazi Jews have.
Sldo, remember that Egypt had always had a white, blue-eyed, blonde/red-haired, demographic component
and that all the Arabs in Egypt are later Invaders.
A scientific team genetic study of human historic evolution has just published a broad study in UK, lead by the German scientist Wolfgang Haak. The team consisted of at least 39 international scientific experts from all over Eurasia.
Some new finding are absolutely revolutionary and stunning for everone interested in such studies.
“Ecology and evolution”
“The genetic history of admixture across inner Eurasia”
https://www.nature.com/articles/s41559-019-0878-2
Now, some of the oldest lies begin to crumble peace by peace. These scientist speak truth, not in any form diminished or deformed by any form of political correctness. This is also the real reason, why such studies – allthough lead by the German team of 3 – are not allowe to be publicised in Germany.
For people who understand German, here is the only publicly known Person – Ingo Bading – who careflly studied all the new findings of evolutionare biology (genetic) over a long time now:
https://www.youtube.com/watch?v=ffqVANXdITQ
Some tips are also given, how to obtain these important papers without paying a fair amount of money – from a special Russian website.
I think Lithuania is a quite interesting case. Some linguistic said, that Lithuanian is the oldest form of indo-European languages. On the other hand Lithuanian DNA is very close to Finnobaltics, which speak their own language from Ural. So are the Lithuanians bilingual with several thousand years of experience.
Thanks for this Detlef. This is an amazing field which has countless facets that people are studying around the clock, for good and not so good reasons.
Political correctness also demands hockey stick climate model in order to tax the poor. Several degrees higher average temperature would and did melt the ice sheet of Arctic Ocean and that would give the water highway from Kola pennisula to east to the rivers of Russia and all the way to China, and the opposite way up. The first silk road.
It might be interesting to know how the conclusion was reached that Abraham was a turkish man.
“and other people may find that they are close to Abraham, a Turkish man (E1b1) who led a group of Anatolians to what he must too have felt was the promised land.”
Thanks Gary, but I think we are just beginning to peel the onion on this.
Comments are closed.